Features
FISH-quant was designed to help you in the analysis of smFISH images.
Python
Analysis is performed with Python.
Open source
Entire code hosted publically on GitHub.
Scalable
Modular workflow suited for high-content applications.
Python
Analysis is performed with Python.
Open source
Entire code hosted publically on GitHub.
Scalable
Modular workflow suited for high-content applications.
Packages
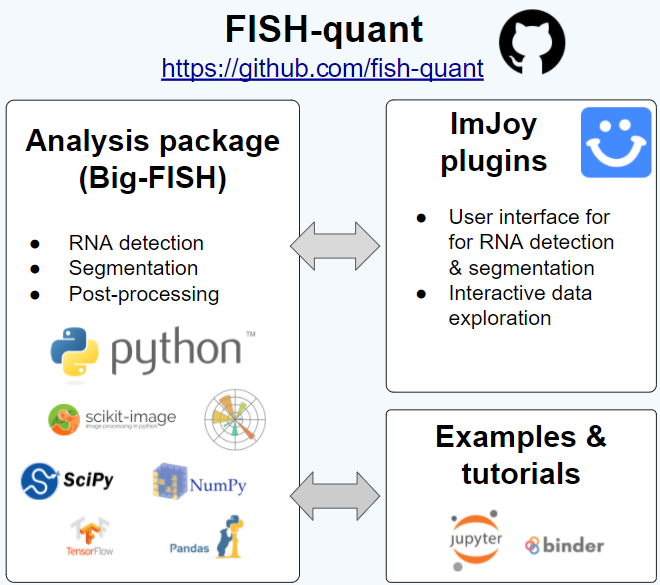
We provide several packages for different analysis tasks and levels of experience. For a detailed description of FISH-quant, we invited you to read our paper.
big-fish allows you to custom build your own analysis pipeline. The Jupyter noteboook tutorials explain the different analysis modules. To get a first feeling, you can start them on binder (starting the remote server can take some time): Blog
January 2022
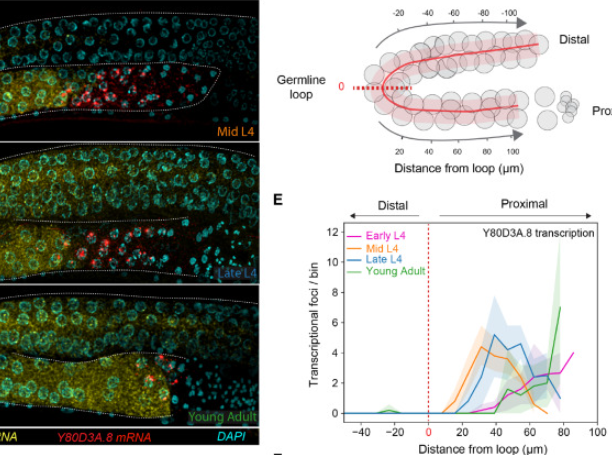
We contributed a FISH-quant analysis to a study looking at transcriptional silencing in C. elegans Developmental Cell.
March 2021
Our paper describing FISH-quant is available on Bioarxiv.
March 2021
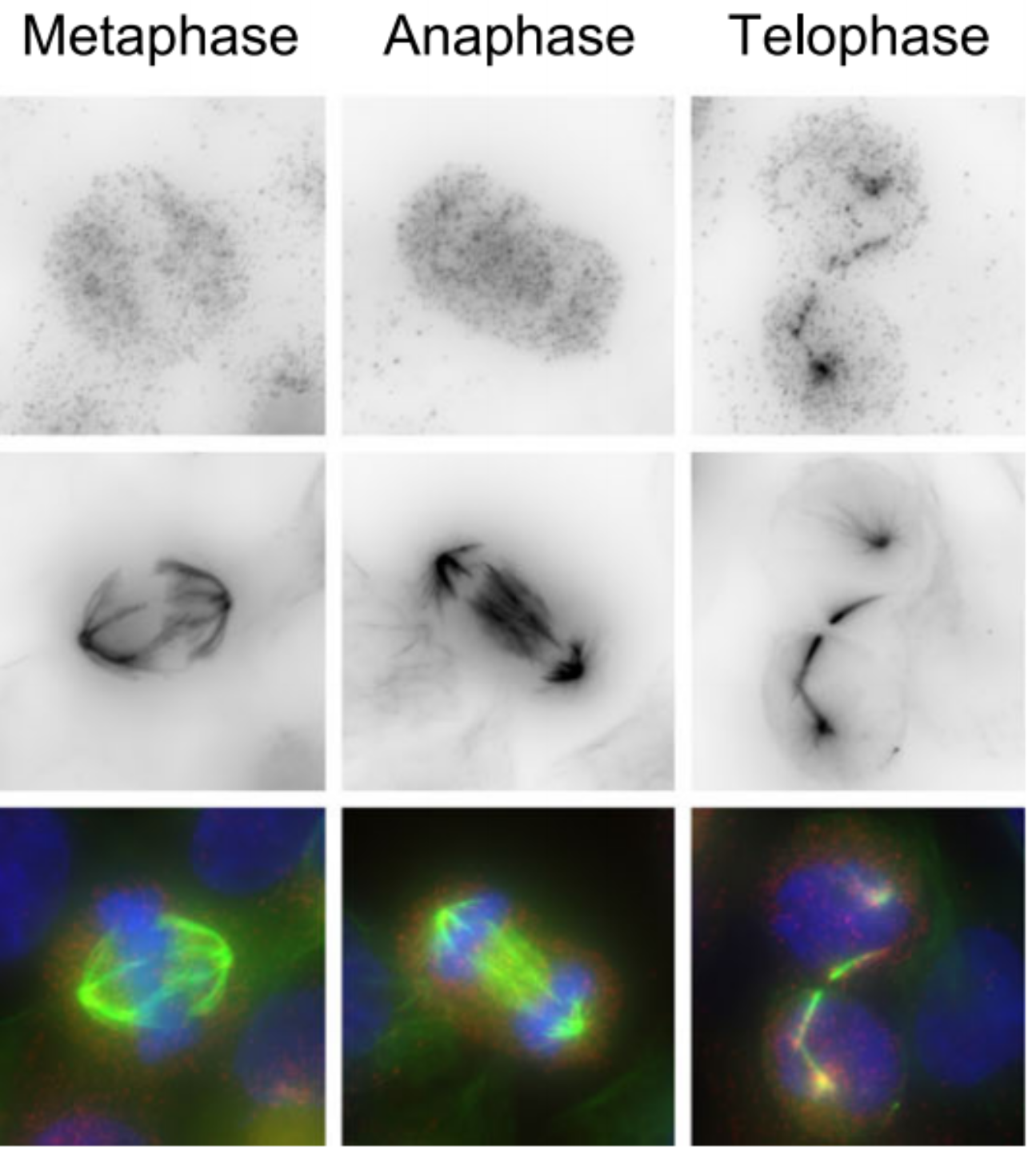
Our paper studying local translation at centrosomes was published in Nature Communications.
September 2020
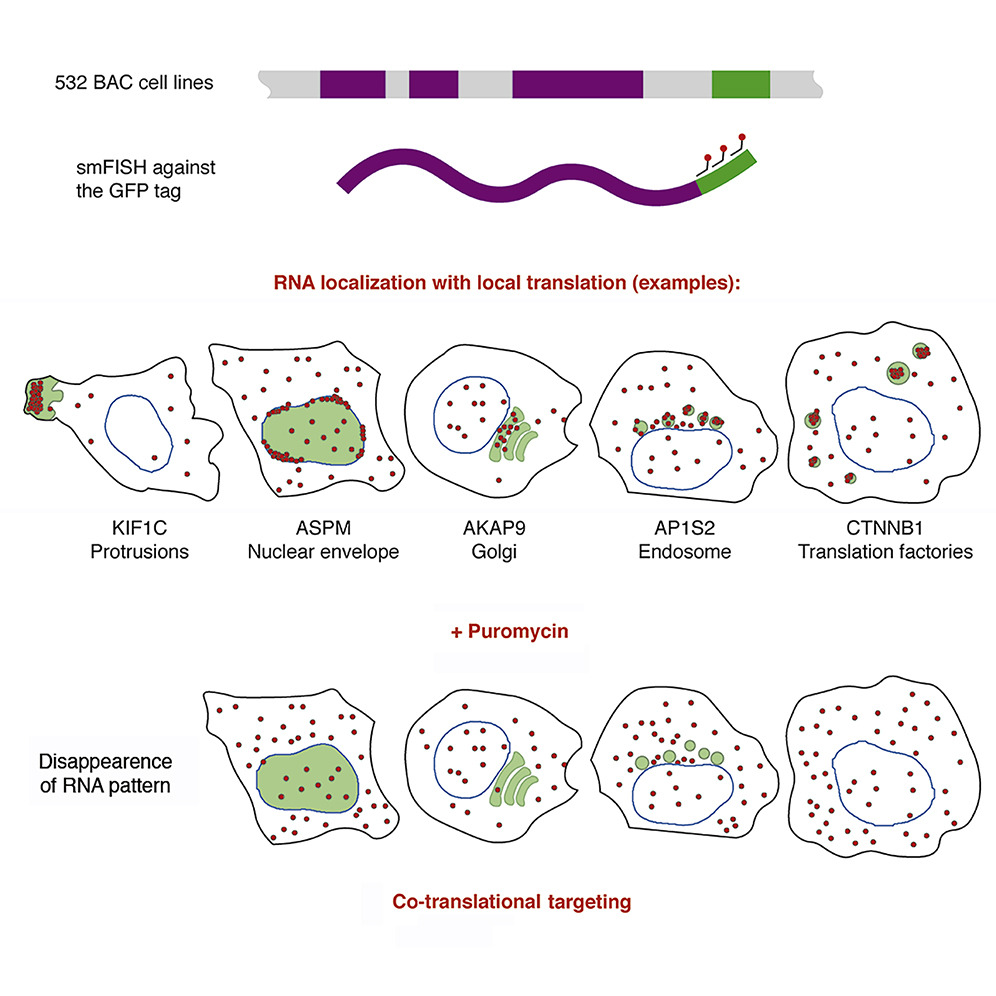
Our paper linking RNA localization and local translation is published in Developmental Cell. To get an overview, you can also read the nice preview from Chin & Lecuyer.

Lead Developer

Project supervision, Developer

Project supervision, Developer

Developer
Contact Us
If you encounter a bug or you have a suggestion for an improvement, please file a GitHub issue in the respective repository.
For any other question, don’t hesitate to contact us by email
Florian Mueller: muellerf.reserach@gmail.com
Arthur Imbert: arthur.imbert@mines-paristech.fr
Thomas Walter: thomas.walter@mines-paristech.fr